Ever wished a picture could have more depth? Researchers at Washington University (WU) in St. Louis have developed a machine learning algorithm capable of doing just that.
A machine learning algorithm developed at the McKelvey School of Engineering at WU can create a continuous 3D structure starting from flat, 2D pictures. So far, the system has been used to create models of human cells from a set of partial images taken using standard microscopy tools in use today.
Such a system could greatly improve the ease with which researchers can interpret 3-dimensional structures from 2D pictures, with potential applications in fields ranging from material science to medicine.
Un-flattening
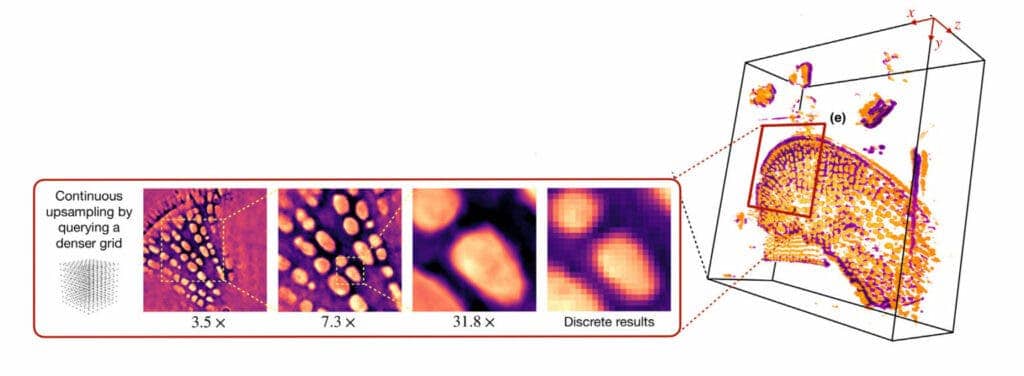
“We train the model on the set of digital images to obtain a continuous representation,” said Ulugbek Kamilov, assistant professor of electrical and systems engineering and of computer science and engineering. “Now, I can show it any way I want. I can zoom in smoothly and there is no pixelation.”
The algorithm is based on a neural field network, a particular machine-learning architecture with the ability to link physical traits to particular spatial coordinates. After receiving adequate training, such a system can be asked to interpret what it’s seeing at any location in an image. Neural field networks were favored as the basis for the new algorithm as they require relatively lower amounts of data for training compared to other types of neural networks. They can be reliably employed to analyze an image as long as sufficient 2D samples are provided for the analysis, the team explains.
For this particular algorithm, the network was trained using standard microscopy images. They consisted of optical pictures of cells: the samples were lit from below, with the light passing through them being recorded.
These images were then fed through the network together with information regarding the internal structure of the cell at particular points of the images. From those, the network was asked to recreate the overall structure of the cells it was being shown. Its output was then judged for how well it reflected reality in a reiterative process.
Once the system’s predictions adequately matched real-life measurements, the network was considered ready. At this point, it was asked to fill in the parts of the cell’s model that could not be captured through direct measurements in the lab.
The team explains that one of the advantages of their new algorithm is that it removes the need for laboratories to maintain data-dense image galleries of any cells they are working with, as a complete representation of its structure can be recreated whenever needed by the network. It also allows for the model to be manipulated in ways that a photograph could never allow.
“I can put any coordinate in and generate that view,” Kamilov says. “Or I can generate entirely new views from different angles.” He can use the model to spin a cell like a top or zoom in for a closer look; use the model to do other numerical tasks; or even feed it into another algorithm.”
The paper “Recovery of continuous 3D refractive index maps from discrete intensity-only measurements using neural fields” has been published in the journal Nature Machine Intelligence.
Was this helpful?



